This package allows plots to be managed in a browser or RStudio’s viewer. The operations that can be managed include saving plots in formats such as png and svg, zooming plots, etc.
Package version is 1.3.1. Checked with R version 4.2.2.
Install Package
Run the following command. Compilation is required during installation.
#Install Package
install.packages("httpgd")Example
See the command and package help for details.
#Loading the library
library("httpgd")
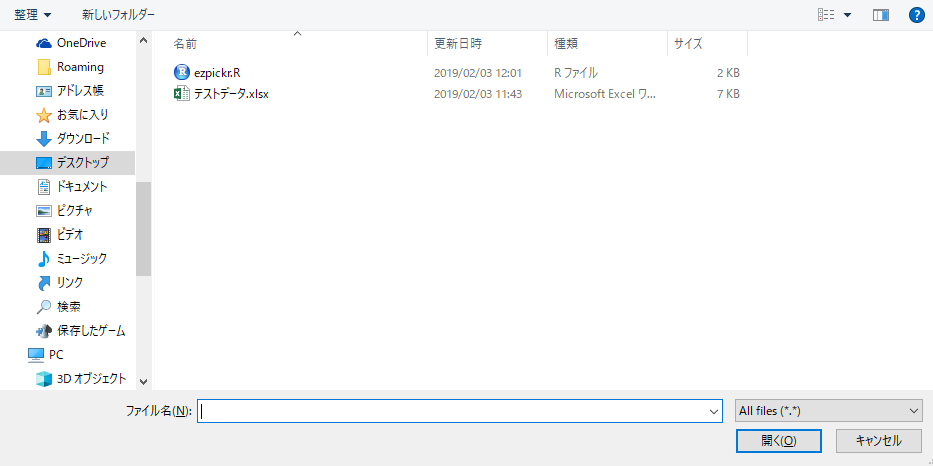
#Preparing to display the plot in the browser: hgd command
#Works with your system's default browser
#Plots to be executed after this command are displayed
hgd()
#Viewing plots in default browser: hgd_browse command
hgd_browse()
#Viewing plots in RStudio: hgd_view command
hgd_view()
#Unlink the default browser: hgd_close command
hgd_close()
#Show httpgd_graphics_device information: hgd_info command
hgd_info(which = dev.cur())
#############################################
###Example: Viewed in a default browser######
#############################################
hgd()
###Creating Data#####
#Install the tidyverse package if it is not already present
if(!require("tidyverse", quietly = TRUE)){
install.packages("tidyverse");require("tidyverse")
}
set.seed(1234)
n <- 300
TestData <- tibble(Group = sample(paste0("Group", 1:4), n,
replace = TRUE),
X_num_Data = sample(c(1:50), n, replace = TRUE),
Y_num_Data = sample(c(51:100), n, replace = TRUE),
Chr_Data = sample(c("か", "ら", "だ", "に",
"い", "い", "も", "の"),
n, replace = TRUE),
Fct_Data = factor(sample(c("か", "ら", "だ", "に",
"い", "い", "も", "の"),
n, replace = TRUE)))
#######
#Install the GGally package if it is not already present
if(!require("GGally", quietly = TRUE)){
install.packages("GGally");require("GGally")
}
#Plotting data features at once: GGally::ggpairs command
ggpairs(data = TestData, columns = c(1, 5, 2, 3),
mapping = aes(color = Group),
upper = list(continuous = "smooth"),
lower = list(combo = "facetdensity"),
diag = list(continuous = "barDiag"),
cardinality_threshold = 30)
#Plotting multiple graphs: GGally:: command
PlotList <- list()
list(for (i in 1:3) {
#Box plot
PlotList[[i]] <- qplot(data = TestData, x = Group,
y = X_num_Data, fill = Group, geom = "boxplot")
#Scatter plot
PlotList[[i + 3]] <- qplot(data = TestData, x = Y_num_Data,
y = X_num_Data, color = Group, geom = "point") +
ggtitle("TEST")
})
#Plot
ggmatrix(PlotList, nrow = 2, ncol = 3,
xAxisLabels = 1:3, yAxisLabels = 1:2, title = "TEST")
#Create ggplot2 objyect
One_Cotinuous <- ggplot(TestData, aes(x = X_num_Data,
color = Group,
fill = Group))
#geom_area command
One_Cotinuous +
geom_area(stat = "count", alpha = 0.7) +
scale_fill_manual(values = c("#a87963", "#505457",
"#4b61ba", "#A9A9A9")) +
labs(title = "geom_area")
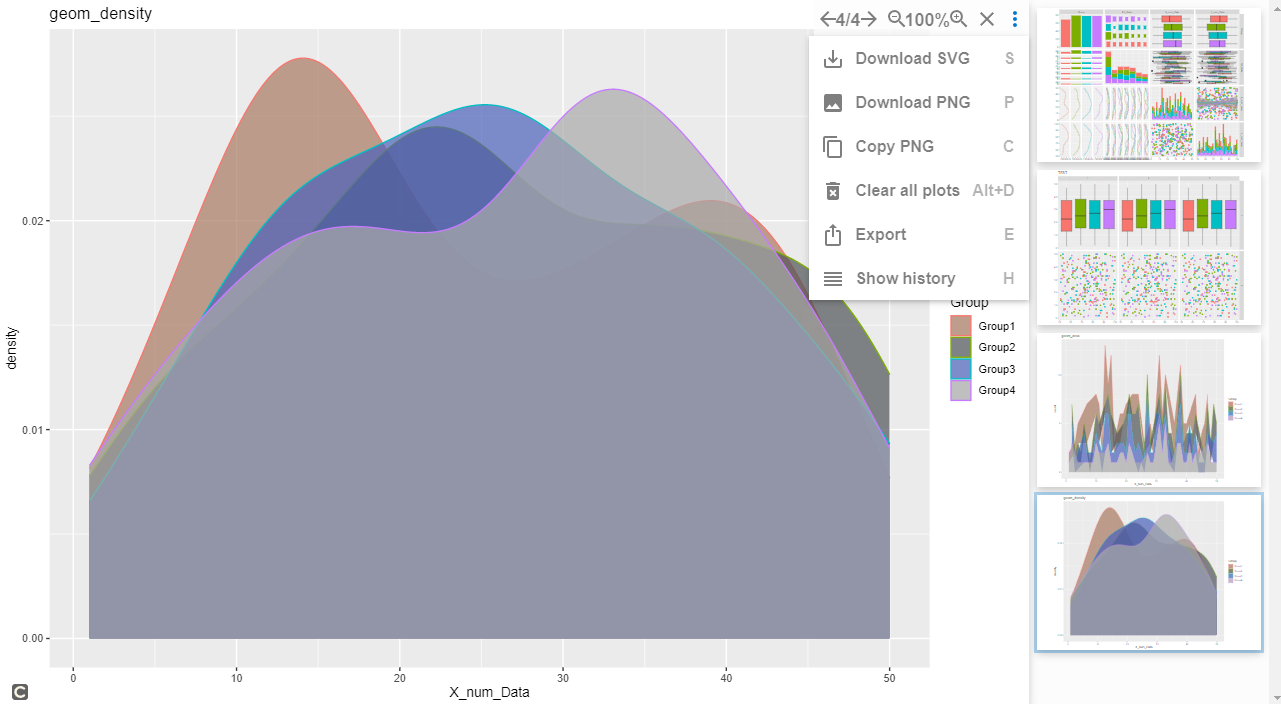
#geom_density command
One_Cotinuous +
geom_density(alpha = 0.7) +
scale_fill_manual(values = c("#a87963", "#505457",
"#4b61ba", "#A9A9A9")) +
labs(title = "geom_density")
#Viewing plots in default browser: hgd_browse command
hgd_browse()
########Output Example
I hope this makes your analysis a little easier !!